Research Interests
Protein phosphorylation. Quality control of High-throughput phosphoproteomic data and statistical analysis.
Protein phosphorylation is the most abundant post-translational modification and a very attractive area of research for understanding biological complexity, cell regulation, for therapeutics, diagnostics and even for synthetic biology. By mutating phosphorylation sites (p-sites), molecular pathways and phenotypes may be manipulated. High-throughput (HTP) Phosphoproteomics has revolutionized this field, especially in the last five years. Despite the flood of these HTP data, there are concerns about their quality and about the level of biological noise in them. This project aims to address the issue of ‘noise’, provide highly curated and annotated datasets (in a database), use them for investigating the general properties, mechanisms and evolution of phosphorylation and develop relevant computational tools.
Estimating the total number of phosphoproteins and phosphorylation sites in eukaryotic proteomes.
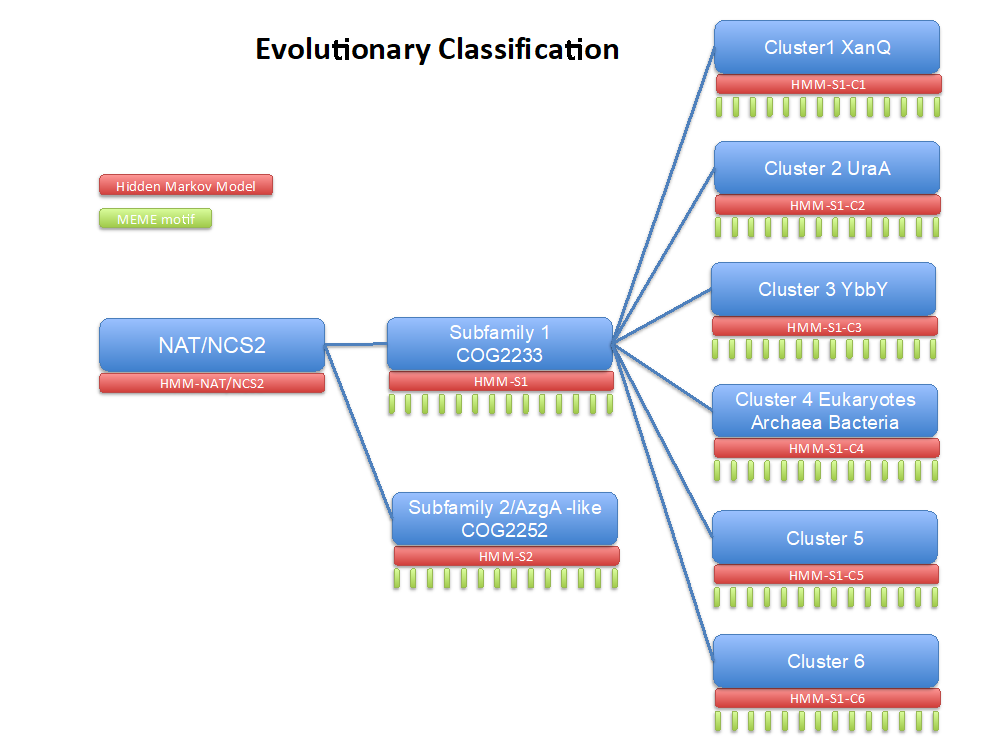
NAT/NCS2 nucleobase transporters
Nucleobase transporters are members of the APC superfamily of amino acid & nucleotide transporters (Saier et al, 2016) and are very important for the cells since their substrate uptake may function as nitrogen/carbon source, affect energy production, replication and protein synthesis through the salvage pathways for nucleotide synthesis (Papakostas et al, 2013; King et al, 2006). In addition to their important direct role on the central metabolism of the cell, they have been regarded as potentially important drug targets for viral/parasitic diseases, for cancer (Köse & Schiedel, 2009) or even mammalian physiology (sleep, drug and alcohol abuse, pain), since they regulate the levels of free nucleosides in circulation.
There exist 5 known families of transporters that use nucleobases as their main substrate, but the NAT/NCS2 family distinguishes from the others because it is very conserved and wide-spread across many different taxa (Frillingos, 2012).
Based on an evolutionary and bioinformatics analysis pipeline that we have already implemented in another superfamily, the tRNA synthetases – see (Chaliotis et al. 2017), we have generated a hierarchical evolutionary classification of the NAT/NCS2 family for prokaryotes. This classification helps to assign family members to evolutionary groups with distinct substrate preferences and identify key conserved and functional amino acids that affect substrate specificity.
Bacterial genomics
The last two decades the field of bacterial genomics has been revolutionized by the development of high-throughput sequencing technologies (both short- and long- read). This wealth of genomic data has enabled the conduct of comparative genomic studies, which aim to identify the core, accessory and species-specific genomic elements of a certain bacterial species. The core genome is defined as the set of genes that are present in all strains of a species, while the accessory genome is a set that is present only in a subset of the strains of that species. The species-specific fingerprints are the genes that are present in all the members of a species and are absent from all the other analyzed strains of the genus. These fingerprints are implicated in the unique lifestyle of a species and can be used for the development of species-specific diagnostic tools. Furthermore, the availability of a large number of bacterial genomes, allows for more accurate species demarcation and delineation of the evolutionary relationships among strains and species of a certain bacterial genus using phylogenomics. Phylogenomics utilize hundreds or even thousands of orthologous genes/proteins shared by all analysed strains and provide higher resolution than the traditional methods based on the 16S rRNA or a few genes (mlst).
- Link to bacterial genomics Web Page
- 05/04/2017: Prof Efstathios Frillingos, School of Medicine, University of Ioannina, Greece. Relatioships between structure, function and substrate specificity of nucleobase transporters.
- 15/06/2016: Prof. Steve Oliver, Dept. of Biochemistry, University of Cambridge, UK. Computational models for Synthetic Biology.
- 03/06/2015: Dr. Liana Tsiatsiani, Utrecht University, Netherlands. Proteolytic enzymes and shotgun proteomics: a win-win situation.
- 20/05/2015: Dr. Eva Fernandez, Research Centre in Evolutionary Anthropology and Paleoecology, Liverpool John Moores University, UK. Ancient human DNA from the Fertile Crescent: reconstructing the primary routes of the Neolithic expansion into Europe.
- 22/04/2015: Dr. Stratos Stratikos, Demokritos Research Center, Athens, Greece. Intracellular antigen processing as a node of adaptive immune response regulation.
- 18/03/2015. Dr. Ioannis Iliopoulos, Dept of Medicine, University of Crete, Greece. Protein Association Discovery in Biomedical Literature.
- 04/04/2012: Prof. Steve Oliver, Dept. of Biochemistry, University of Cambridge. Systems Biology: A science in flux.
- Prof. Steve Oliver, Dept. of Biochemistry, University of Cambridge, UK. Sabbatical visit. 6 - 17 June 2016
- Nikolaidis M, Hesketh A, Frangou N, Mossialos D, Van de Peer Y, Oliver SG, Amoutzias GD. A panoramic view of the genomic landscape of the genus Streptomyces. In press: Microbial Genomics.
- Goutzourelas N, Kevrekidis DP, Barda S, Malea P, Trachana V, Savvidi S, Kevrekidou A, Assimopoulou AN, Goutas A, Liu M, Lin X, Kollatos N, Amoutzias GD, Stagos D. Antioxidant Activity and Inhibition of Liver Cancer Cells' Growth of Extracts from 14 Marine Macroalgae Species of the Mediterranean Sea. Foods. 2023 Mar 19;12(6):1310. doi: 10.3390/foods12061310. PMID: 36981236; PMCID:PMC10048654.
- Yfantis A, Mylonis I, Chachami G, Nikolaidis M, Amoutzias GD, Paraskeva E, Simos G. Transcriptional Response to Hypoxia: The Role of HIF-1-Associated Co-Regulators. Cells. 2023 Mar 3;12(5):798. doi: 10.3390/cells12050798. PMID:36899934; PMCID: PMC10001186.
- Papanikolaou NA, Nikolaidis M, Amoutzias GD, Fouza A, Papaioannou M, Pandey A, Papavassiliou AG. The Dynamic and Crucial Role of the Arginine Methylproteome in Myoblast Cell Differentiation. Int. J. Mol. Sci. 2023, 24(3), 2124; https://doi.org/10.3390/ijms24032124.
- Tsakogiannis D, Nikolaidis M, Zagouri F, Zografos E, Kottaridi C, Kyriakopoulou Z, Tzioga L, Markoulatos P, Amoutzias GD, Bletsa G. Mutation Profile of HPV16 L1 and L2 Genes in Different Geographic Areas. Viruses 2022 Dec 31;15(1):141. doi: 10.3390/v15010141.
- Georgi I, Asoutis Didaras N, Nikolaidis M, Dimitriou TG, Charistos L, Hatjina F, Amoutzias GD, Mossialos D. The Impact of Vairimorpha (Nosema) ceranae Natural Infection on Honey Bee (Apis mellifera) and Bee Bread Microbiota. Applied Sciences (Switzerland), 2022, 12(22), 11476.
- Baltsavia I, Theodosiou T, Papanikolaou N, Pavlopoulos GA, Amoutzias GD, Panagopoulou M, Chatzaki E, Andreakos E, IIliopoulos I. Prediction and Ranking of Biomarkers Using multiple UniReD. Int J Mol Sci. 2022 Sep 21;23(19):11112. doi: 10.3390/ijms231911112
- Nikolaidis M, Hesketh A., Mossialos D. Iliopoulos I. Oliver S.G., Amoutzias G.D. A Comparative Analysis of the Core Proteomes within and among the Bacillus subtilis and Bacillus cereus Evolutionary Groups Reveals the Patterns of Lineage- and Species-Specific Adaptations. Microorganisms 2022, 10(9), 1720; doi: 10.3390/microorganisms10091720.
- Amoutzias GD, Nikolaidis M, Hesketh A. The Notable Achievements and the Prospects of Bacterial Pathogen Genomics. Microorganisms. 2022 May 17;10(5):1040. doi: 10.3390/microorganisms10051040.
- Nikolaidis M, Papakyriakou A, Chlichlia K, Markoulatos P, Oliver SG, Amoutzias GD. Comparative Analysis of SARS-CoV-2 Variants of Concern, Including Omicron, Highlights Their Common and Distinctive Amino Acid Substitution Patterns, Especially at the Spike ORF. Viruses. 2022 Mar 29;14(4):707. doi: 10.3390/v14040707.
-
Nikolaidis M, Markoulatos P, Van de Peer Y, Oliver SG,
Amoutzias GD. The Neighborhood of the Spike Gene Is a
Hotspot for Modular Intertypic Homologous and Nonhomologous
Recombination in Coronavirus Genomes. Mol Biol Evol. 2022 Jan
7;39(1):msab292. doi: 10.1093/molbev/msab292.
Mentioned in the Forbes Magazine by William Haseltine - Amoutzias GD, Nikolaidis M, Tryfonopoulou E, Chlichlia K, Markoulatos P, Oliver SG. The Remarkable Evolutionary Plasticity of Coronaviruses by Mutation and Recombination: Insights for the COVID-19 Pandemic and the Future Evolutionary Paths of SARS-CoV-2. Viruses. 2022 Jan 2;14(1):78. doi: 10.3390/v14010078.
- Bletsa G, Zagouri F, Amoutzias GD, Nikolaidis M, Zografos E, Markoulatos P, Tsakogiannis D. Genetic variability of the HPV16 early genes and LCR. Present and future perspectives. Expert Rev Mol Med. 2021 Dec 1;23:e19. doi: 10.1017/erm.2021.18.
- Nikolaidis M, Tsakogiannis D, Bletsa G, Mossialos D, Kottaridi C, Iliopoulos I, Markoulatos P, Amoutzias GD. HPV16-genotyper: A computational tool for risk-assessment, lineage genotyping and recombination detection in hpv16 sequences, based on a large-scale evolutionary analysis. Diversity Volume 13, Issue 10 October 2021 Article number 497
- Koureas M, Amoutzias GD, Vontas A, Kyritsi M, Pinaka O, Papakonstantinou A, Dadouli K, Hatzinikou M, Koutsolioutsou A, Mouchtouri VA, Speletas M, Tsiodras S, Hadjichristodoulou C. Wastewater monitoring as a supplementary surveillance tool for capturing SARS-COV-2 community spread. A case study in two Greek municipalities. Environ Res. 2021 Sep;200:111749. doi: 10.1016/j.envres.2021.111749.
- Theodorakis E, Antonakis AN, Baltsavia I, Pavlopoulos GA, Samiotaki M, Amoutzias GD, Theodosiou T, Acuto O, Efstathiou G, Iliopoulos I. ProteoSign v2: a faster and evolved user-friendly online tool for statistical analyses of differential proteomics. Nucleic Acids Res. 2021 Jul 2;49(W1):W573-W577. doi: 10.1093/nar/gkab329.
- Tsadila C, Nikolaidis M, Dimitriou TG, Kafantaris I, Amoutzias GD, Pournaras S, Mossialos D. Antibacterial Activity and Characterization of Bacteria Isolated from Diverse Types of Greek Honey against Nosocomial and Foodborne Pathogens. Applied Sciences (Switzerland), Volume 11, Issue 131 July 2021 Article number 5801.
- Didaras NA, Kafantaris I, Dimitriou TG, Mitsagga C, Karatasou K, Giavasis I, Stagos D, Amoutzias GD, Hatjina F, Mossialos D. Biological Properties of Bee Bread Collected from Apiaries Located across Greece. Antibiotics (Basel). 2021 May 10;10(5):555. doi: 10.3390/antibiotics10050555.
- Kafantaris I, Tsadila C, Nikolaidis M, Tsavea E, Dimitriou TG, Iliopoulos I, Amoutzias GD, Mossialos D. Transcriptomic Analysis of Pseudomonas aeruginosa Response to Pine Honey via RNA Sequencing Indicates Multiple Mechanisms of Antibacterial Activity. Foods. 2021 Apr 24;10(5):936. doi: 10.3390/foods10050936.
- Koutsianos D, Athanasiou LV, Dimitriou T, Nikolaidis M, Tsadila C, Amoutzias G, Mossialos D, Koutoulis KC. Antibiotic Resistance Patterns and mcr-1 Detection in Avian Pathogenic Escherichia coli Isolates from Commercial Layer and Layer Breeder Flocks Demonstrating Colibacillosis in Greece. Microb Drug Resist. 2020 Sep 21. doi: 10.1089/mdr.2020.0057.
- Daskou M, Tsakogiannis D, Alexopoulou DS, Dimitriou TG, Mossialos D, Amoutzias GD, Kottaridi C, Markoulatos P. A colorimetric IsoPCR for the rapid and sensitive visual detection of high-risk HPV16 in clinical samples with hydroxynaphthol blue. J Virol Methods. 2021 Apr;290:114072. doi: 10.1016/j.jviromet.2021.114072
- Kafantaris, I., Amoutzias, G.D. & Mossialos, D. Foodomics in bee product research: a systematic literature review. Eur Food Res Technol (2020). https://doi.org/10.1007/s00217-020-03634-5
- Samaras A, Nikolaidis M, Antequera-Gómez ML, Cámara-Almirón J, Romero D, Moschakis T, Amoutzias GD, Karaoglanidis GS. Whole Genome Sequencing and Root Colonization Studies Reveal Novel Insights in the Biocontrol Potential and Growth Promotion by Bacillus subtilis MBI 600 on Cucumber. Front Microbiol. 2021 Jan 12;11:600393. doi: 10.3389/fmicb.2020.600393.
- Koureas M, Kalompatsios D, Amoutzias GD, Hadjichristodoulou C, Gourgoulianis K, Tsakalof A. Comparison of Targeted and Untargeted Approaches in Breath Analysis for the Discrimination of Lung Cancer from Benign Pulmonary Diseases and Healthy Persons. Molecules. 2021 Apr 29;26(9):2609.doi: 10.3390/molecules26092609.
- Daskou M, Dimitriou TG, Alexopoulou DS, Tsakogiannis D, Amoutzias GD, Mossialos D, Kyriakopoulou Z, Markoulatos P. WarmStart colorimetric RT-LAMP for the rapid, sensitive and specific detection of Enteroviruses A-D targeting the 5'UTR region. J Appl Microbiol. 2021 Jan;130(1):292-301. doi: 10.1111/jam.14770.
- Didaras NA, Karatasou K, Dimitriou TG, Amoutzias GD, Mossialos D. Antimicrobial Activity of Bee-Collected Pollen and Beebread: State of the Art and Future Perspectives. Antibiotics (Basel). 2020 Nov 14;9(11):811. doi: 10.3390/antibiotics9110811.
- Koureas M, Kirgou P, Amoutzias G, Hadjichristodoulou C, Gourgoulianis K, Tsakalof A. Target Analysis of Volatile Organic Compounds in Exhaled Breath for Lung Cancer Discrimination from Other Pulmonary Diseases and Healthy Persons. Metabolites. 2020 Aug 3;10(8):317. doi: 10.3390/metabo10080317.
- Nikolaidis M, Mossialos D, Oliver SG, Amoutzias GD. Comparative Analysis of the Core Proteomes among the Pseudomonas Major Evolutionary Groups Reveals Species-Specific Adaptations for Pseudomonas aeruginosa and Pseudomonas chlororaphis. Diversity 2020, 12(8), 289; https://doi.org/10.3390/d12080289
- Komaitis F, Kalliampakou K, Botou M, Nikolaidis M, Kalloniati C, Skliros D, Du B, Rennenberg H, Amoutzias GD, Frillingos S, Flemetakis E. Molecular and physiological characterization of the monosaccharide transporters gene family in Medicago truncatula. J Exp Bot. 2020 May 30;71(10):3110-3125. doi: 10.1093/jxb/eraa055.
- Daskou M, Dimitriou TG, Kouklamani-Giannouli G, Nikolaidis M, Mossialos D, Amoutzias GD, Markoulatos P. Development of a reverse transcription loop-mediated isothermal amplification assay (RT-LAMP) that detects enteroviruses by targeting the highly conserved 5'-UTR region. Virus Genes. 2020 Apr;56(2):194-201. doi: 10.1007/s11262-020-01732-w. Epub 2020 Jan 18. PMID: 31955385
- Theodosiou T, Papanikolaou N, Savvaki M, Bonetto G, Maxouri S, Fakoureli E, Eliopoulos AG, Tavernarakis N, Amoutzias GD, Pavlopoulos GA, Aivaliotis M, Nikoletopoulou V, Tzamarias D, Karagogeos D, Iliopoulos I. UniProt-Related Documents (UniReD): assisting wet lab biologists in their quest on finding novel counterparts in a protein network. NAR Genomics and Bioinformatics, 2020 1–13. doi: 10.1093/nargab/lqaa005.
- Ntountoumi C, Vlastaridis P, Mossialos D, Stathopoulos C, Iliopoulos I, Promponas V, Oliver SG, Amoutzias GD. Low complexity regions in the proteins of prokaryotes perform important functional roles and are highly conserved. Nucleic Acids Res. 2019 Sep 4. pii: gkz730. doi: 10.1093/nar/gkz730.
- Daskou M, Tsakogiannis D, Dimitriou TG, Amoutzias GD, Mossialos D, Kottaridi C, Gartzonika C, Markoulatos P. WarmStart colorimetric LAMP for the specific and rapid detection of HPV16 and HPV18 DNA. J Virol Methods. 2019 Apr 28;270:87-94.
- Daskou M, Tsakogiannis D, Dimitriou TG, Manali M, Apti C, Amoutzias GD, Mossialos D, Kottaridi C, Markoulatos P. Α 2-stage, nested-like nucleic acid amplification method (IsoPCR) for the highly sensitive detection of HPV16 and HPV18 DNA. Mol Cell Probes. 2019 Mar 19. pii: S0890-8508(18)30307-4.
- Nikolaidis M, Mimouli K, Kyriakopoulou Z, Tsimpidis M, Tsakogiannis D, Markoulatos P, Amoutzias GD. Large-scale genomic analysis reveals recurrent patterns of intertypic recombination in human enteroviruses. Virology 2019 Jan 2;526:72-80.
- Chaliotis A, Vlastaridis P, Ntountoumi, C, Botou M, Yalelis V, Lazou P, Tatsaki E, Mossialos D, Frillingos S and Amoutzias GD. NAT/NCS2-hound: A webserver for the detection and evolutionary classification of prokaryotic and eukaryotic nucleobase-cation symporters of the NAT/NCS2 family. Gigascience. 2018 Dec 1;7(12). doi: 10.1093/gigascience/giy133.
- Karakasidou K, Nikolouli K, Amoutzias GD, Pournou A, Manassis C, Tsiamis G,Mossialos D. Microbial diversity in biodeteriorated Greek historical documents dating back to the 19th and 20th century: A case study. In press, Microbiology Open, 2018. DOI: 10.1002/mbo3.596
- Priftis A, Goutzourelas N, Chalabalaki M, Ntasi G, Stagos D, Amoutzias GD, Skaltsounis LA, Kouretas D. Effect of polyphenols from coffee and grape on gene expression in myoblasts. Mech Ageing Dev. 2017 Nov 23. pii: S0047-6374(17)30155-0. doi: 10.1016/j.mad.2017.11.015.
- Tsakogiannis D, Moschonas GD, Bella E, Kyriakopoulou Z, Amoutzias GD, Dimitriou TG, Kottaridi C, Markoulatos P. Association of p16 (CDKN2A) polymorphisms with the development of HPV16-related precancerous lesions and cervical cancer in the Greek population. J Med Virol. 2017 Nov 23. doi: 10.1002/jmv.24996.
- Kyriakopoulou Z, Amoutzias GD, Dimitriou TG, Tsakogiannis D, Mossialos D, Markoulatos P. Intra- and inter-serotypic recombinations in the 5΄ UTR-VP4 region of Echovirus 30 strains. Arch Virol. 2017 Oct 30. doi: 10.1007/s00705-017-3600-1.
- Fikatas A, Dimitriou TG, Kyriakopoulou Z, Moschonas GD, Amoutzias GD, Mossialos D, Gartzonika C, Levidiotou-Stefanou S, Markoulatos P. Detection of negative and positive RNA strand of poliovirus Sabin 1 and echovirus E19 by a stem-loop reverse transcription PCR. Lett Appl Microbiol. 2017 Jun 20. doi: 10.1111/lam.12766. PMID: 28631392
- Vlastaridis P, Kyriakidou P, Chaliotis A, Van de Peer Y, Oliver SG, Amoutzias GD. Estimating the total number of phosphoproteins and phosphorylation sites in eukaryotic proteomes. Gigascience. 2017 Feb 1;6(2):1-11. doi: 10.1093/gigascience/giw015. PMID:28327990
- Vlastaridis P., Papakyriakou A., Chaliotis A., Stratikos E., Oliver SG. and Amoutzias GD. The pivotal role of protein phosphorylation in the control of yeast central metabolism. G3: Genes, Genomes, Genetics 2017 Mar 1. pii: g3.116.037218. PMID: 28250014
- Tsimpidis M, Bachoumis G, Mimouli K, Kuriakopoulou Z, Robertson DL, Markoulatos P, Amoutzias GD.T-RECs: Rapid and large-scale detection of recombination events among different evolutionary lineages of viral genomes. BMC Bioinformatics. 2017 Jan 5;18(1):13. PMID:28056784
- Chaliotis A, Vlastaridis P, Mossialos D, Ibba M, Becker HD, Stathopoulos C and Amoutzias GD. The complex evolutionary history of aminoacyl-tRNA synthetases. Nucleic Acids Res. 2017 Feb 17;45(3):1059-1068. PMID: 28180287
- Fikatas A, Dimitriou TG, Kyriakopoulou Z, Tsachouridou O, Gartzonika C, Levidiotou-Stefanou S, Amoutzias GD, Markoulatos P. Serum neutralization assay for the determination of antibody levels against non-polio enterovirus strains in Central and Western Greece. Viral Immunol. 2016 Sep;29(7):444-50. PMID: 27410516
- Amoutzias GD, Giannoulis T, Moutou KA, Psarra AG, Stamatis C, Tsipourlianos A, Mamuris Z. SNP identification through transcriptome analysis of the European brown hare (Lepus europaeus): cellular energetics and mother’s curse. PLoS One. 2016 Jul 26;11(7). PMID: 27459096
- Vlastaridis P, Oliver SG, Van de Peer Y, Amoutzias GD. The Challenges of Interpreting Phosphoproteomics Data: A Critical View Through the Bioinformatics Lens. Lecture Notes in Bioinformatics. 2016. 9874, pp. 196-204.
- Amoutzias GD, Chaliotis A, Mossialos D. Discovery Strategies of Bioactive Compounds Synthesized by Nonribosomal Peptide Synthetases and Type-I Polyketide Synthases Derived from Marine Microbiomes. Mar Drugs. 2016 Apr 16;14(4). pii: E80. PMID: 27092515
- Pappi PG, Maliogka VI, Amoutzias GD, Katis NI. Genetic variation of eggplant mottled dwarf virus from annual and perennial plant hosts. Arch Virol. 2016 Mar;161(3):631-9. PMID: 26660163
- Katsiani, A.T., Maliogka, V.I., Amoutzias, GD, Efthimiou, K.E., Katis, N.I. Insights into the genetic diversity and evolution of Little cherry virus 1. Plant Pathology 2015, 64 (4), pp. 817-824. DOI: 10.1111/ppa.12309
- Kyriakopoulou Z, Bletsa M, Tsakogiannis D, Dimitriou TG, Amoutzias GD, Gartzonika C, Levidiotou-Stefanou S, Markoulatos P. Molecular epidemiology and evolutionary dynamics of Echovirus 3 serotype. Infect Genet Evol. 2015 Jun;32:305-12. PMID: 25791929
- Kyriakopoulou Z, Pliaka V, Amoutzias GD, Markoulatos P. Recombination among human non-polio enteroviruses: implications for epidemiology and evolution. Virus Genes. 2015 Apr;50(2):177-88 PMID: 25537948
- Tsakogiannis D, Kyriakopoulou Z, Ruether IG, Amoutzias GD, Dimitriou TG, Diamantidou V, Kotsovassilis C, Markoulatos P. Determination of HPV16 physical status through E1/E6 and E2/E6 ratio analysis. J Med Microbiol. 2014 Sep 11. pii: jmm.0.076810-0. PMID: 25212758
- Ruether IG, Dimitriou TG, Tsakogiannis D, Kyriakopoulou Z, Amoutzias GD, Gartzonika C, Levidiotou-Stefanou S, Markoulatos P. Characterization of novel intergenogroup and intergenotype recombinant noroviruses from central Greece. Mol Cell Probes. 2014 Aug;28(4):204-10. PMID: 24751495
- Tsakogiannis D, Darmis F, Gortsilas P, Ruether IG, Kyriakopoulou Z, Dimitriou TG, Amoutzias G, Markoulatos P. Nucleotide polymorphisms of the human papillomavirus 16 E1 gene. Arch Virol. 2014 Jan;159(1):51-63. PMID: 23881083
- Tsakogiannis D, Kyriakopoulou Z, Amoutzias G, Ruether IG, Dimitriou TG, Panotopoulou E, Markoulatos P. Identification of novel E6-E7 sequence variants of human papillomavirus 16. Arch Virol. 2013 Apr;158(4):821-8. PMID: 23208280
- Stagos D, Amoutzias GD, Matakos A, Spyrou A, Tsatsakis AM, Kouretas D. Chemoprevention of liver cancer by plant polyphenols. Food Chem Toxicol. 2012 Jun;50(6):2155-70. PMID: 22521445
- Cock MJ, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, et al. The Ectocarpus Genome and Brown Algal Genomics. The Ectocarpus Genome Consortium. Advances in Botanical Research. 2012. Volume 64, 2012, Pages 141-184. DOI: 10.1016/B978-0-12-391499-6.00005-0
- Amoutzias GD, He Y, Lilley KS, Van de Peer Y, Oliver SG. Evaluation and properties of the budding yeast phosphoproteome. Mol Cell Proteomics. 2012 Jun;11(6):M111.009555. PMID: 22286756
- Cock MJ, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010 Jun 3;465(7298):617-21. PMID: 20520714
- Amoutzias GD, He Y, Gordon J, Mossialos D, Oliver SG, Van de Peer Y. Posttranslational regulation impacts the fate of duplicated genes. Proc Natl Acad Sci U S A. 2010 Feb 16;107(7):2967-71. PMID: 20080574
- Mossialos D, Amoutzias GD. Role of siderophores in cystic fibrosis (CF) pathogenesis: foes or friends? Int J Med Microbiol. 2009 Feb;299(2):87-98. PMID: 18755629
- Amoutzias GD, Robertson DL, Van de Peer Y, Oliver SG. Choose your partners: Dimerization in eukaryotic transcription factors. Trends Biochem Sci. 2008 May;33(5):220-9. PMID: 18406148
- Amoutzias GD, Van de Peer Y, Mossialos D. Evolution and taxonomic distribution of non-ribosomal peptide and polyketide synthases. Future Microbiol. 2008 Jun;3:361-70. PMID: 18505401
- Amoutzias G, Van de Peer Y. Together we stand: genes cluster to coordinate regulation. Dev Cell. 2008 May;14(5):640-2. PMID: 18477446
- Pinney JW, Amoutzias GD, Rattray M, Robertson DL. Reconstruction of ancestral protein interaction networks for the bZIP transcription factors. Proc Natl Acad Sci U S A. 2007 Dec 18;104(51):20449-53. PMID: 18077348
- Pampalakis G, Arampatzidou M, Amoutzias G, Kossida S, Sotiropoulou G. Identification and analysis of mammalian KLK6 orthologue genes for prediction of physiological substrates. Comput Biol Chem. 2007 Nov 29. PMID: 18243805
- Mossialos D, Amoutzias G. Siderophores in fluorescent pseudomonads: new tricks from an old dog. Future Microbiol. 2007 Aug;2:387-95. PMID: 17683275
- Amoutzias GD, Pichler EE, Mian N, De Graaf D, Imsiridou A, Robinson-Rechavi M, Bornberg-Bauer E, Robertson DL, Oliver SG. A protein interaction atlas for the nuclear receptors: Properties and quality of a hub-based dimerisation network. BMC Syst Biol. 2007 Jul 31;1(1):34. PMID: 17672894
- Holden BJ, Pinney JW, Lovell SC, Amoutzias GD & Robertson DL. An exploration of alternative visualizations of the basic helix-loop-helix protein interaction network. BMC Bioinformatics. 2007 Aug 6;8(1):289. PMID: 17683601
- Amoutzias G, Veron A, Weiner A, Robinson-Rechavi M, Bornberg-Bauer E, Oliver S, Robertson D. One Billion Years of bZIP Transcription Factor Evolution: Conservation and Change in Dimerization, and DNA-Binding Site Specificity. Mol Biol Evol. 2006 Dec 28. PMID: 17194801
- Amoutzias GD, Bornberg-Bauer E, Oliver SG, Robertson DL. The Reduction/oxidation-phosphorylation control of the DNA binding of the bZIP family is linked with its dimerization network. BMC Genomics 2006 May 4;7:107. PMID:
- Podowski RM, Cleary JG, Goncharoff NT, Amoutzias G, Hayes WS. Suregene, a scalable system for automated term disambiguation of gene and protein names. J Bioinform Comput Biol. 2005 Jun;3(3):743-70. PMID: 16108092
- Amoutzias GD, Weiner J, Bornberg-Bauer E. Phylogenetic profiling of protein interaction networks in eukaryotic transcription factors reveals focal proteins being ancestral to hubs. Gene. 2005 Feb 23. PMID: 15777629
-
Amoutzias GD, Robertson DL, Oliver SG, Bornberg-Bauer E.
Convergent evolution of gene networks by single-gene
duplications in higher eukaryotes.. EMBO Rep. 2004
Mar;5(3):274-9. PMID:
14968135
Commented in: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1299035/
Awarded: http://documents.manchester.ac.uk/display.aspx?DocID=7475 - Amoutzias GD, Robertson DL, Bornberg-Bauer E. The evolution of protein interaction networks in regulatory proteins. Comp Funct Genomics 2004; 5: 79-84. PMID: 18629034
- Podowski RM, Cleary JG, Goncharoff NT, Amoutzias G, Hayes WS. AZuRE, a scalable system for automated term disambiguation of gene and protein names. Proc IEEE Comput Syst Bioinform Conf. 2004;:415-24. PMID: 16448034
- Imsiridou A, Hardy H, Maudling N, Amoutzias G, Zaldivar Comenges JM. Web database of molecular genetic data from fish stocks. J Hered. 2003 May-Jun;94(3):265-7. PMID: 12816969
Evolution of the tRNA-synthetases superfamily.
Aminoacyl-tRNA synthetases (AARSs) mediate the accurate esterification of amino acids to their cognate tRNAs and thus represent an essential superfamily of enzymes responsible for the fidelity of the genetic code. Interestingly, gene duplication, divergence and gene loss are much more frequent events than originally considered. Paralogues or even paralogue fragments of AARSs exhibit diverse functions outside translation, spanning from editing and antibiotic resistance in bacteria to molecular hubs within essential signaling pathways that regulate tumorigenesis in human. Our work focuses on developing new computational skills for identifying these synthetases in proteomes, on storing the data in a database scheme and on analyzing data from thousands of prokaryotic genomes in order to understand what drives the evolution and diversity of this superfamily.
Link to web application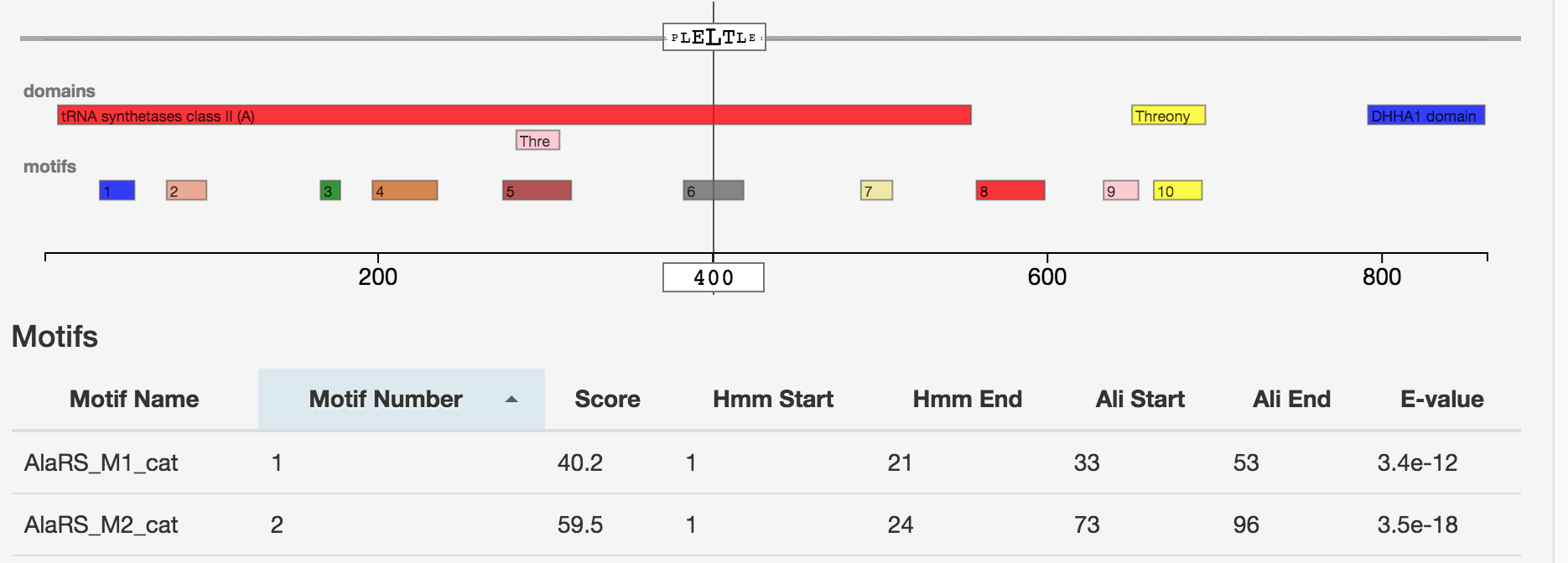
Evolution of gene regulation with a special interest on protein interaction networks and dimerization of transcription factors (bZIP, bHLH, Nuclear Receptors).
In many eukaryotic transcription factor gene families, proteins require a physical interaction with an identical molecule or with another molecule within the same family to form a functional dimer and bind DNA. Depending on the choice of partner and the cellular context, each dimer triggers a sequence of regulatory events that lead to a particular cellular fate, for example, proliferation or differentiation. Recent syntheses of genomic and functional data reveal that partner choice is not random; instead, dimerization specificities, which are strongly linked to the evolution of the protein family, apply. Our focus is on understanding these interaction specificities, their functional consequences and how they evolved. This knowledge is essential for understanding gene regulation and designing a new generation of drugs.
Research Grants awarded
Filtering, Annotation and Bioinformatics analyses of high-throughput Phosphoproteomic data (FAB-PHOS). Principal Investigator: GD Amoutzias. Funded by Aristeia II - GSRT.
Bioinformatic analysis of human genomic and phosphoproteomic
data. Principal Investigator: GD Amoutzias.
Funded by The Research Committee of the University of Thessaly.
Collaborations
Due to the interdisciplinary nature of Bioinformatics, the lab has a series of ongoing collaborations with experimental as well as in-silico labs such as:
Prof. Markoulatos and Dr. Mosialos, Laboratory of Virology and Bacteriology, Dept. Biochemistry and Biotechnology, University of Thessaly, Greece.
Prof. Van de Peer, Bioinformatics and Genomics Laboratory, Dept. of Plant Systems Biology, VIB/UGent, Belgium.
Prof. Robertson, Viral Genomics and Bioinformatics Laboratory, Centre for Virus Research, Institute of Infection, Immunity and Inflammation, University of Glasgow, UK.
Prof. Oliver, Yeast Systems Biology laboratory, Dept. of Biochemistry, Cambridge University, UK.
Prof. Frillingos, School of Medicine, University of Ioannina, Greece.
Assist. Prof. Papanikolaou, School of Medicine, Aristotle University of Thessaloniki, Greece.
Prof. Stathopoulos, Laboratory of Biochemistry, School of Medicine, University of Patra, Greece.
Prof. Katis and Dr. Maliogka, Laboratory of Plant Pathology, School of Agriculture, Aristotle University of Thessaloniki, Greece
Dr. Stratikos, Demokritos Research Center, Athens Greece.