T-RECs (Tool for RECombinations)

Recombination along with point mutations are the main mechanisms of mutation contributing to virus evolution, especially in RNA viruses, such as HIV, Enteroviruses, Noroviruses etc. Although point mutations allow some investigation of the evolutionary fitness landscape, recombination leads to the virus to perform “jumps” within this evolutionary landscape and explore other regions. Furthermore, recombination has been identified as a mechanism of combining advantageous properties from various genomes into a new one, of eradicating deleterious mutations (i.e. act against Muller’s Ratchet), for development of drug resistance, or for evasion from the immune system. In addition, it has a disruptive effect on molecular phylogenetic analysis. Therefore, it is paramount to be able to detect such events and eventually correlate them with new properties or new outbreaks.
The rapid advances in sequencing technologies in combination with the small size of viral genomes is creating an explosion of genomic data in the public databases. Nevertheless, many of the current recombination tools are limited by the number of sequences being analyzed at one time, or their user-friendliness. Therefore, there is a need for a new generation of tools that will rapidly scan (in a user-friendly environment) hundreds or even thousands of genomes or sequence fragments and detect candidate recombination events that may later be further analyzed with more sensitive and specialized methods.
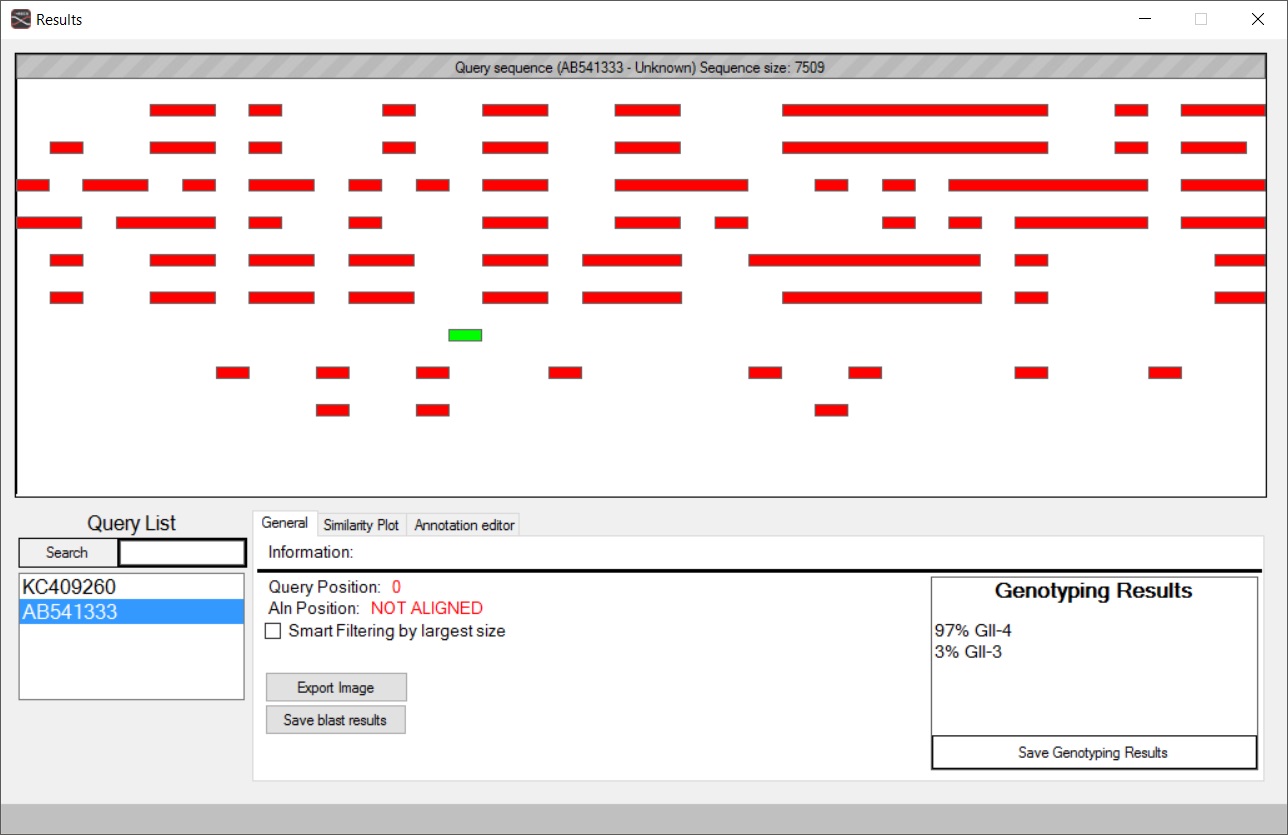
Our motivation was to develop a computational pre-filtering tool, named T-RECs (Tool for RECombinations) that is based on the BLASTN heuristic local pairwise alignment method with sliding windows. The tool expands on the basic principles of the NCBI genotyping web tool and the SWeBlast perl scripts, but as a locally installed program in a Microsoft Windows environment, with a user friendly graphical interface that allows large-scale analyses and visualization. It rapidly scans hundreds or even thousands of query genomes or even sequence fragments, allows genotyping based on a user-defined sequence database and detects candidate recombination events among members of different evolutionary groups e.g. organisms, genogroups, genotypes etc. The detected candidate events may later be further analyzed (within T-RECs) with similarity plots that integrate the Blast results and even genomic/functional/post-translational modification annotations. T-RECs also allows the clustering with Uclust (http://drive5.com/usearch/manual/uclust_algo.html) of sequences based on a user-defined similarity threshold and thus removes redundancy and simplifies large-scale analyses with many highly similar sequences. In addition, the tool allows for certain regions of a sequence to be selected and uploaded to NCBI Blast or saved. After this pre-filtering step with T-RECs, identified targets may be later analyzed with other more specialized methods/tools.
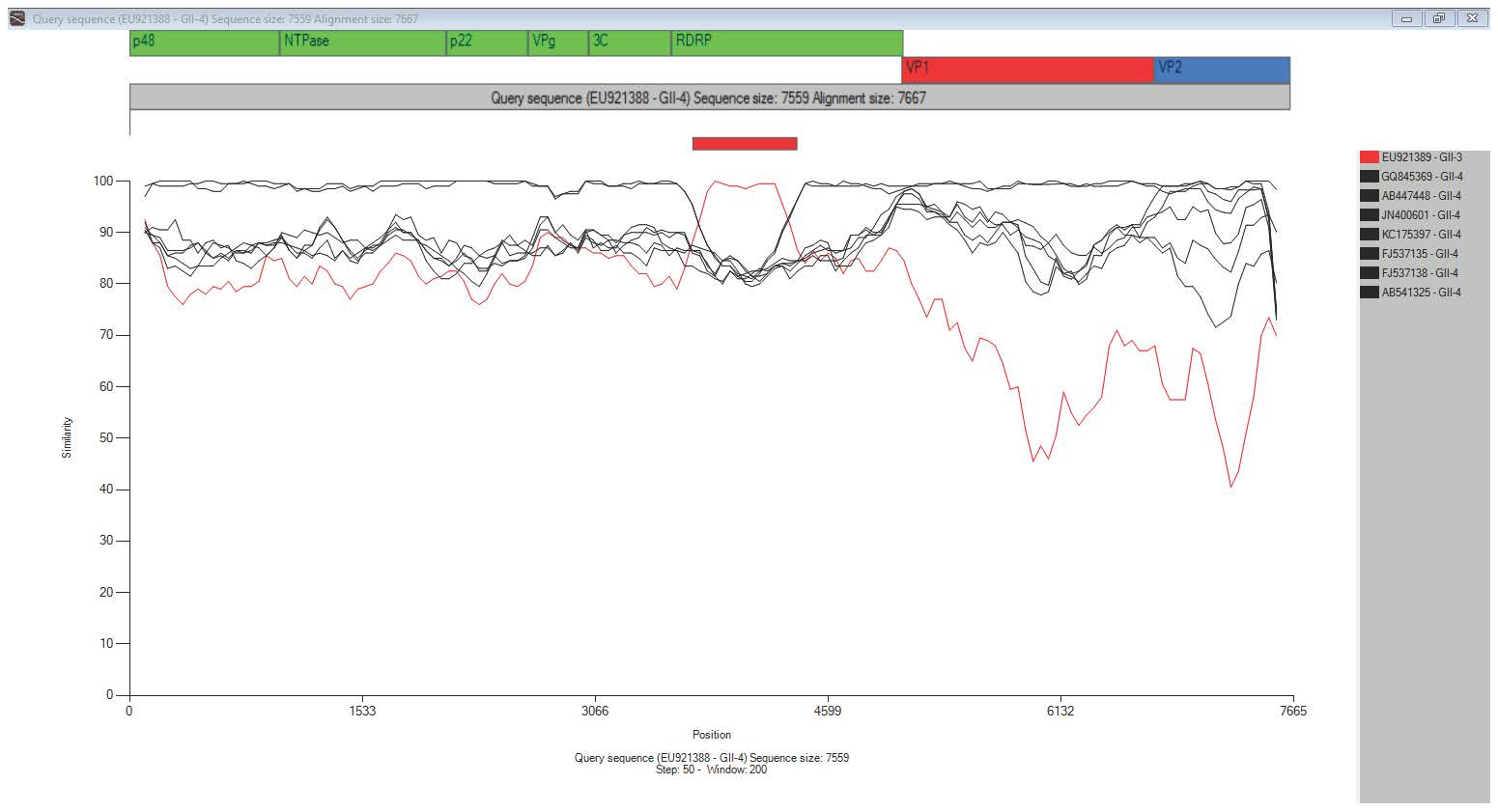
Figure 1. Integrated view of the T-RECs result with similarity plot and ORF annotation.
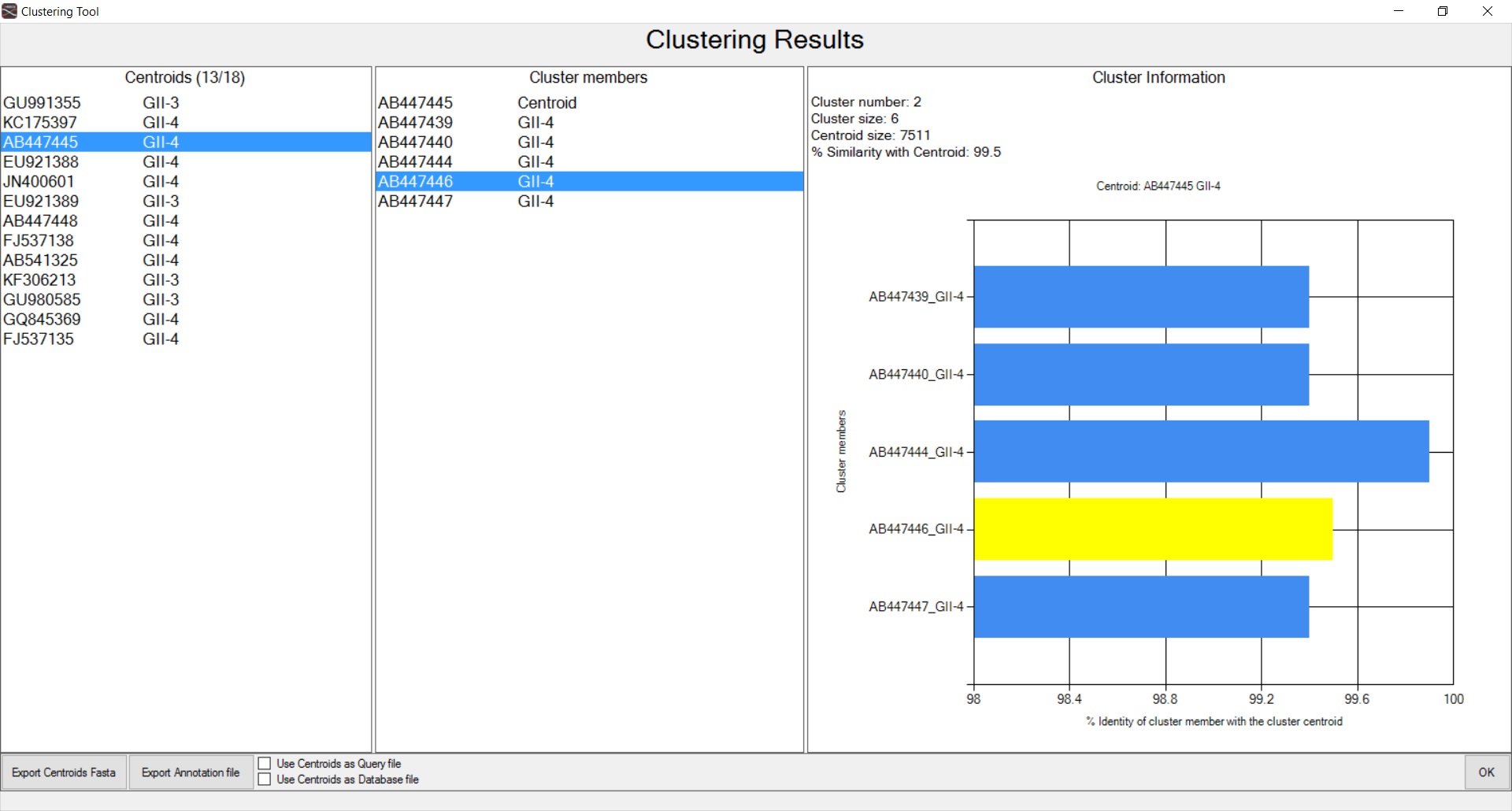
Figure 2. Clustering of sequences, based on nucleotide identity threshold, with Uclust.